Introduction
The discovery of MAPK/ERK pathway mutations has revolutionized the care for patients with histiocytic neoplasms. However, mutational analysis on biopsy specimens is often limited by low tumor cellularity leading to testing failure or disease involvement of difficult-to-reach sites (e.g., central nervous system). Recently, molecular analysis of cell-free DNA (cfDNA) has shown promise as a viable tool for discovering driver mutations in multiple cancers. A recent study of mainly BRAF-mutated non-Langerhans cell histiocytosis (LCH) showed 50% concordance between tumor and plasma cfDNA analysis for BRAF-V600E. However, it is unknown whether cfDNA has a role in the evaluation of cases that harbor mutations beyond BRAF-V600E. We conducted this study to address this unanswered question using a large cohort of individuals with diverse histiocytic neoplasms from a tertiary-care institution.
Methods
We included 40 cases with histiocytic neoplasms with stored plasma and germline DNA in this study. Tissue molecular analysis was conducted using a targeted next generation sequencing (NGS) using commercial assays or whole exome sequencing (WES) using an in-house assay. A custom panel of 226 genes, including exonic and intronic regions, was used for the cfDNA assay in this study. To avoid cfDNA false positives, germline mutations identified from normal DNA sequencing were filtered using a BostonGene ®-developed error reduction algorithm.The total length of the analyzed genomic regions was 1.56 MB. For samples with an input greater than 30 ng, a 0.2% variant allele frequency (VAF) limit of detection (LOD) was used; a 0.4% VAF LOD was used for samples less than 30 ng.
Results
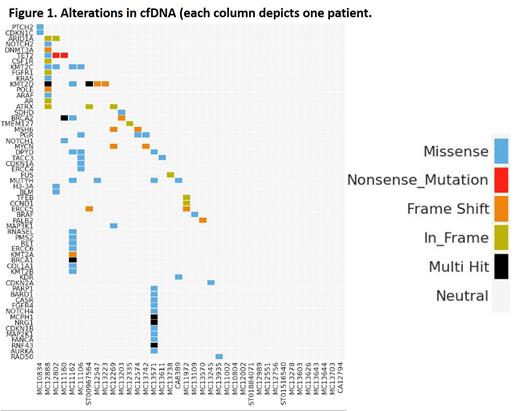
The final analysis cohort included 38 patients with histiocytic neoplasms after exclusion of 2 cases (failure of germline DNA library preparation and mismatch of germline and somatic DNA, respectively). Disease types included Erdheim-Chester disease (ECD; n=13, 34%), Langerhans cell histiocytosis (LCH; n=13, 34%), and Rosai-Dorfman disease (RDD; n=12, 32%). 24 cases were multisystem (63%), and the remaining were single-system. Commonly involved organs included bone (n=25, 66%), kidney (n=12, 32%), skin (n=8, 21%), central nervous system (n=7, 18%), and pituitary (n=7, 18%). Most cases (n=31, 82%) underwent plasma collection pre-treatment, 5 cases underwent collection after histiocytosis-directed therapy, and 2 while on treatment. Tissue NGS/WES data were available for 28 (74%) cases; 5 cases (13%) had insufficient tissue cellularity, and 5 (13%) did not undergo any tissue sequencing. Among those who underwent successful tissue sequencing assays, the most common common driver alterations was BRAF V600E (n=5, 26%). Other alterations included those in MAP2K1 (n=4, 21%), BRAF-fusions (n=3, 10%; partners UBDT2, RNF11, and LMTK2, respectively), and single cases of KRAS, NRAS and BRAF indel, respectively. cfDNA analysis showed at least one variant in 23 cases (60%). Correlation between tissue and cfDNA for known driver mutations was poor and concordant findings were only seen in one case with ECD harboring BRAF V600E in tissue as well as plasma (VAF 0.5%). Among the 5 cases where tissue mutational analysis had failed, cfDNA showed mutations in MUTYH, PTCH2, NOTCH3 SDHD, and TMEM127, respectively (Figure 1). Among the 5 cases where tissue molecular assessment could not be undertaken due to various reasons, cfDNA showed alterations in 4 cases, encompassing NOTCH1, TET2, BRCA1, MSH6, PIK3CD, and FUS genes.
Conclusions
In a large cohort of adults with diverse histiocytic neoplasms, we found poor correlation between tumor and plasma cfDNA molecular findings. The reason for the lack of correlation may be related to low tumor shedding especially among non- BRAF V600E cases. cfDNA analysis may have a role to identify potential drivers of pathogenesis among cases that are unable to successfully undergo tissue molecular analysis. Further studies are underway to characterize other novel alterations that were discovered in the cfDNA analysis.
Disclosures
Goyal:Opna Bio: Membership on an entity's Board of Directors or advisory committees. Efremov:BostonGene: Current Employment, Current equity holder in private company, Current holder of stock options in a privately-held company. Sookiasian:BostonGene: Current Employment, Current equity holder in private company, Current holder of stock options in a privately-held company. Tazearslan:BostonGene: Current Employment, Current equity holder in private company, Current holder of stock options in a privately-held company. Yudina:BostonGene: Current Employment, Current equity holder in private company, Current equity holder in publicly-traded company. Bennani:Affimed: Other: Advisory board; No personal compensation; Secura Bio: Other: Advisory board; No personal compensation; Kymera: Other: Advisory board; No personal compensation; Astellas Pharma: Other: Advisory board; No personal compensation; Acrotech: Other: Advisory board; No personal compensation; Acrotech: Other: Scientific Advisory Committee, No personal compensation . Shah:Celgene: Research Funding; MRKR Therapeutics: Research Funding; Astellas: Research Funding; AbbVie: Research Funding. Witzig:ADC: Membership on an entity's Board of Directors or advisory committees; Salarius Pharma: Membership on an entity's Board of Directors or advisory committees; Karyopharm: Research Funding; Kura Oncology: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal